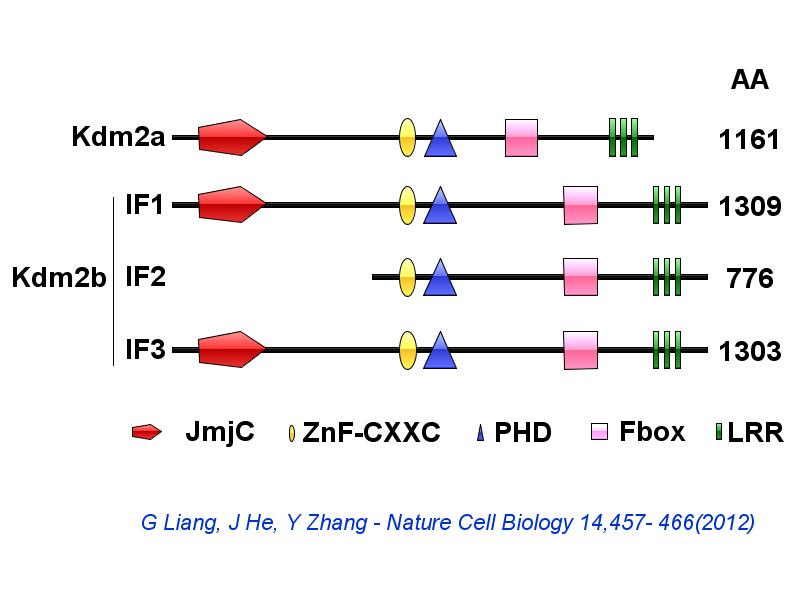
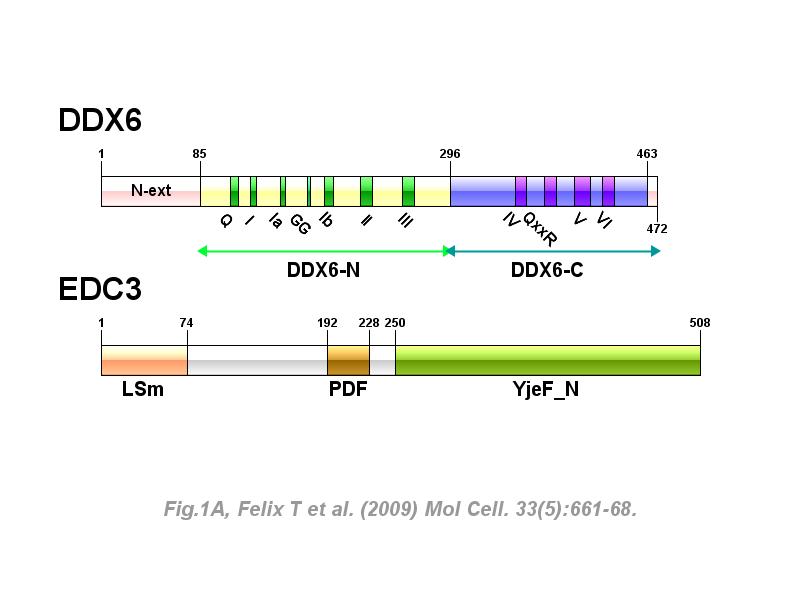
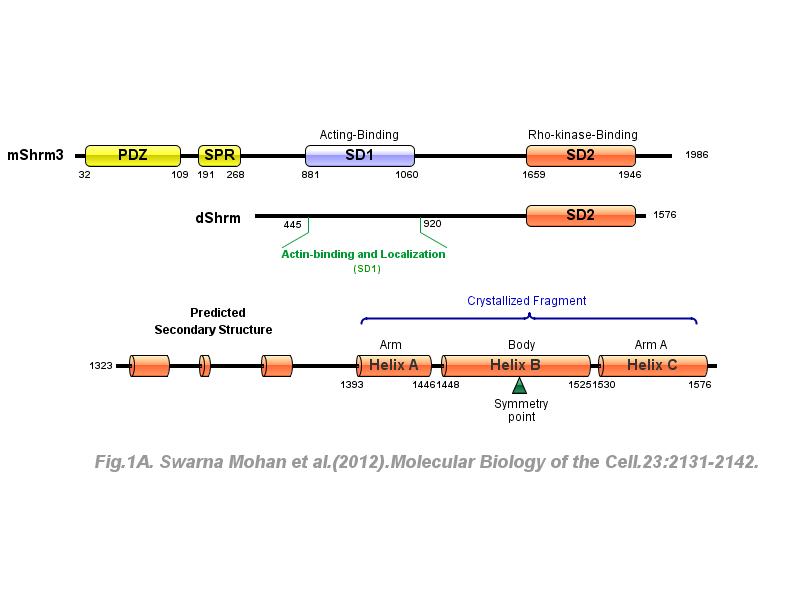
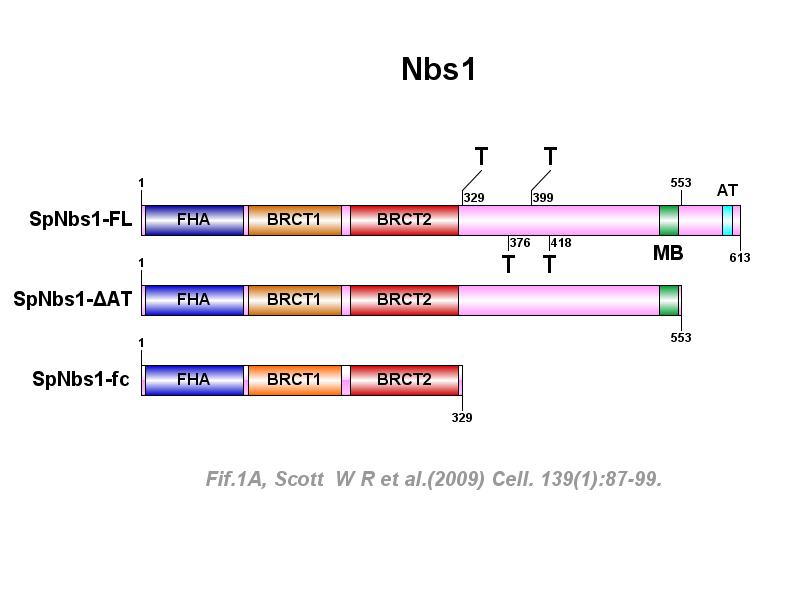
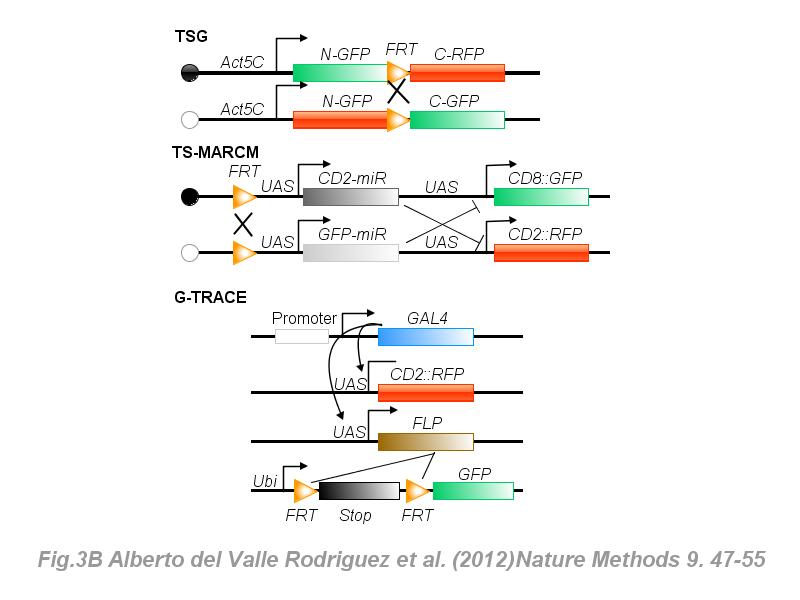
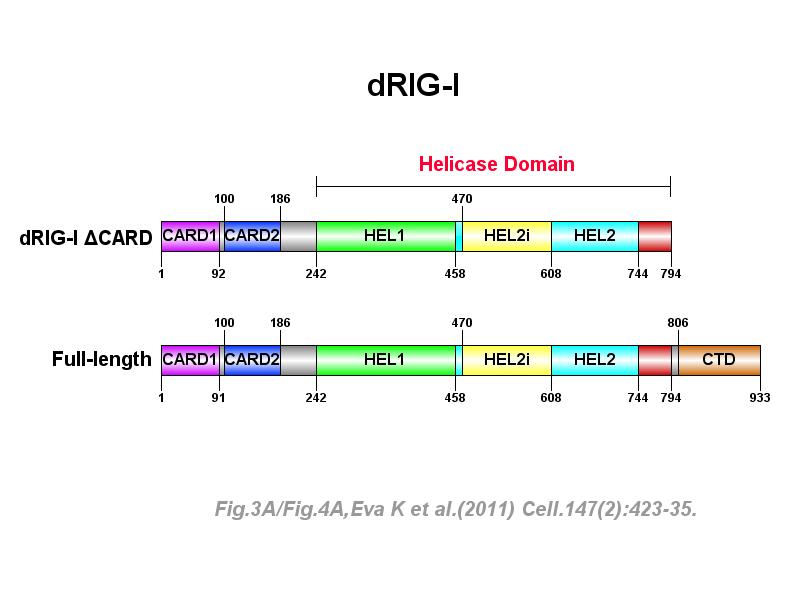
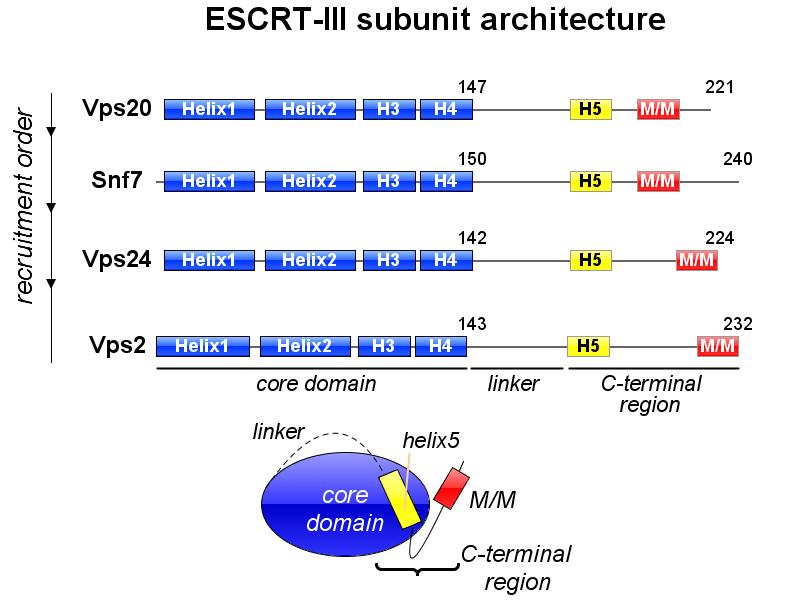
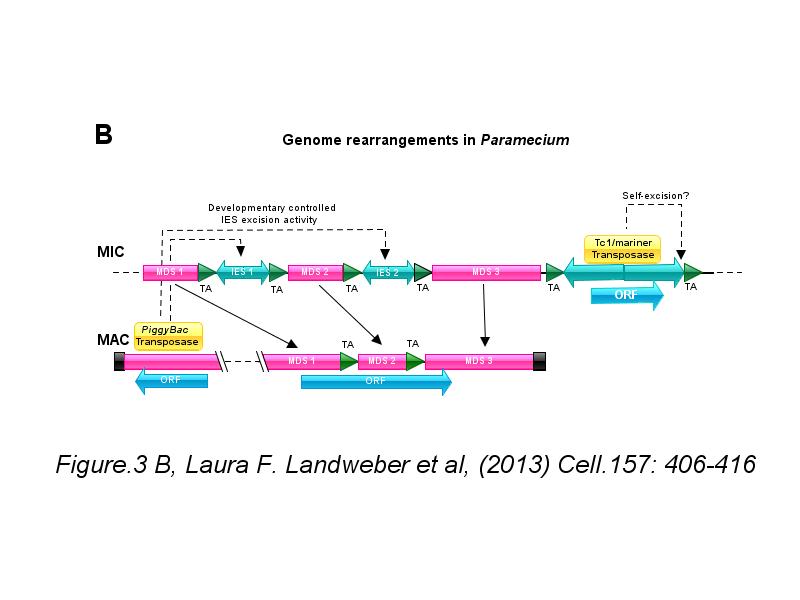
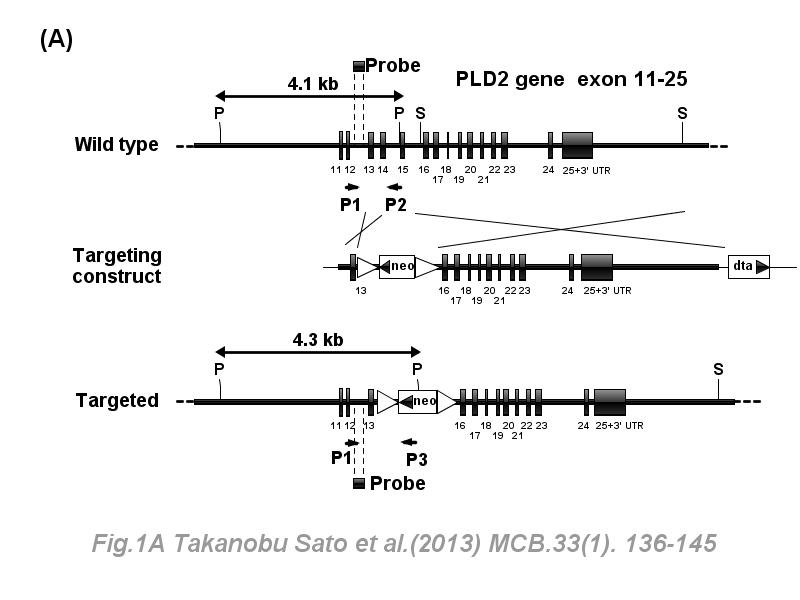
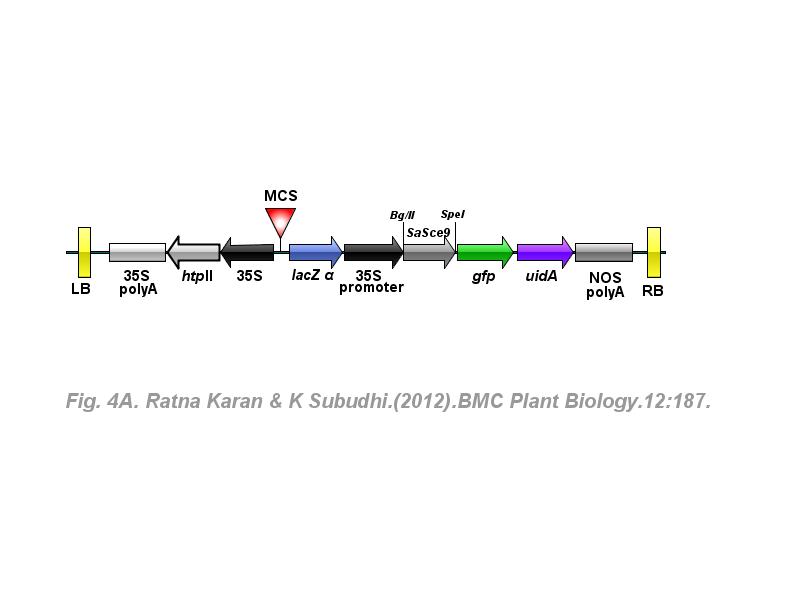
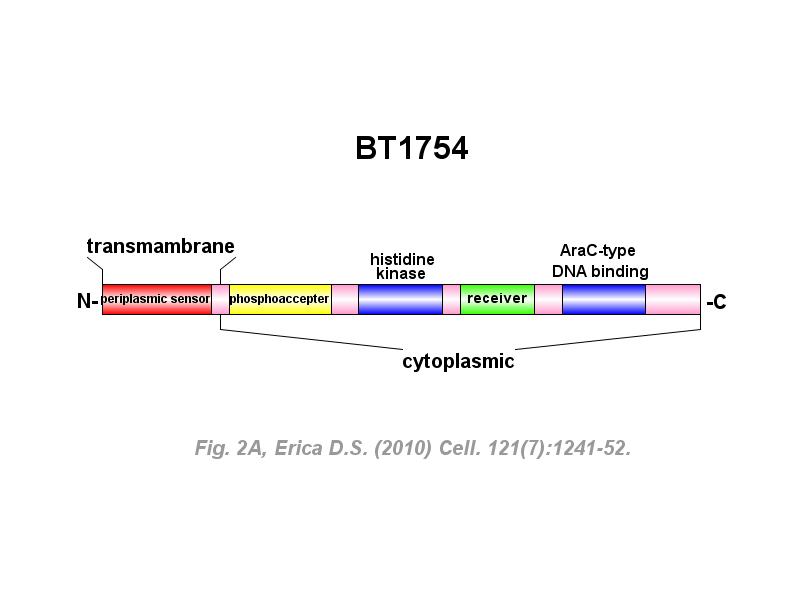
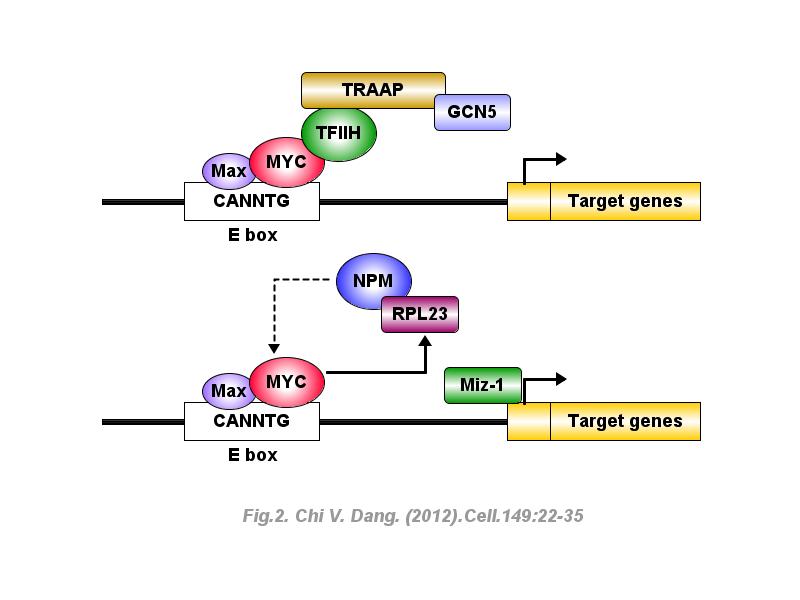
※ IBS INTRODUCTION:
A concise, delicate and precise picture can afford a great advantage over description alone. The graphical representation of biological sequences with various functional elements is fundamental for the efficient introduction and interpretation of scientific findings obtained in the course of the investigation of molecular biological functions. Schematic biological sequence diagrams that enable intuitive and illustrative visualization are not only crucial for the easy understanding of scientific publications, but also for sharing viewpoints on works in progress with colleagues. Therefore, the illustration of biological sequences has become one of the most essential skills for biologists, who at present mainly use the Microsoft PowerPoint, Adobe Illustrator or Photoshop for diagramming biological sequences. However, these software packages were developed for the general purpose of image manipulation, so it is difficult and time consuming to use them to prepare a sequence organization diagram of sufficient accuracy and quality. For example, the scale of functional elements such as DNA motifs and protein domains is typically approximated, even judged by eye. Also, when using these tools, neither the locations nor the positions of functional elements can be precisely designated in the biological sequences.
In 2009, our group developed a software package called Domain Graph (DOG), which can be used for preparing protein domain graphs in a step-by-step manner (Ren et al. 2009).Also, the scales and positions of functional domains and sites can be precisely defined by users. Since publication, DOG has assisted numerous researchers visualizing protein sequence organization. Interestingly, although DOG was specifically designed for protein domain graphs, a number of researchers used it for visualizing the functional elements in nucleotide sequences. From the complete set of citations of DOG, we observed an increasing demand for precisely diagramming functional elements and sites in nucleotide sequences. However, DOG was developed for proteins, so the visualization of nucleotide sequences may not be an optimal application. In this regard, the development of a new software package for the presentation of both protein and nucleotide sequences should prove to be of great help to the research community.
In this work, we present a new tool called Illustrator for Biological Sequences (IBS) for assisting experimentalists in drawing publication-quality diagrams of both protein and nucleotide sequences. With a dual-mode user interface, experimentalists are allowed to produce their own schematic diagrams for either protein or nucleotide sequences in a convenient manner. To provide a user-friendly software package, abundant graphical elements, such as polygons, brackets, curves and polylines, are available for representing functional elements or regulatory molecules. We anticipate this powerful tool will prove to be a great help to scientists for the purpose of visualizing biological sequences.